drraheel@gmail.com

Prof. Dr. Shahid Nadeem Chohan
Prof. Dr. Raheel Qamar
Introduction – Curiosity is an exciting part of human psyche. We are curious about our surroundings as well as ourselves. We want to know what, how and why things are as they are and how can we either use them as such or change them to our advantage. We have been doing this from the beginning of our evolutionary history and keep doing it over our personal life spans. Consequently, this curiosity has led to a tremendous wealth of knowledge. This huge knowledge base has generally been organized in such a way that it potentially can readily be accessed, verified and used. All this organized body of knowledge is collectively called science. Science has several subdivisions; the physical sciences deal with our physical environment and the Biological sciences deal with all life forms including ourselves. While Biology encompasses all sciences dealing with life and its interaction with the environment, Molecular Biology is the study of life at a molecular level. HEC News Letter.
The most important molecules we study in Molecular Biology are two nucleic acids which are RNA (Ribo Nucleic Acid) and DNA (Deoxyribo Nucleic Acid. Out of these two, DNA is more prevalent, present inside each cell and carries the instructions to create and sustain that particular life form. Like a book of life! This is a manual of instructions to create and sustain life no matter whether you are a human, a plant or for that matter a lizard. This may be hard to believe but is true that the whole book of life is written in only four letters. These letters, i.e., A, T, C and G denote four types of nucleotides which respectively are; Adenine, Thymine, Cytosine and Guanine. These nucleotides are the building blocks of DNA which is the hereditary material for almost all life forms. A DNA molecule has a double helical structure composed of two complementary strands of DNA as was discovered by Watson, Crick and Franklin in 1953.

DNA double helix structure.
Image source: www.genecrc.org/site/ lc/lc2b.htm
The sets of instruction in this book of life, individually called genes are arranged as long chains of all four types of nucleotides. These sets of instructions or genes are implemented or expressed in the form of proteins. Like DNA, proteins are also long chains of certain building blocks called amino acids. There are 21 amino acids altogether. Every protein molecule is constituted of one or more types of amino acids but unlike DNA, every building block (i.e., amino acid) is not necessarily represented in every protein. Each protein has different function which ultimately depends on the type of amino acids it comprises of as well as their sequence in it.
The sum of all genes in an organism is called its genome and the sum of all proteins in an organism is called its proteome. Likewise, the study of genomes is called genomics and the study of proteomes is termed as proteomics. The secret of the nature and functioning of a species is encoded in its genes as genome and expressed as its particular proteome. The genome and therefore the proteome of one species is different from the same of another and that is the basis of the speciation. For example genome and therefore genes of a human are different from those of, for example, a horse and so are their products, i.e., proteins. This is what put them apart. Genome of a species is in fact a blue print of its nature and the functioning potentials. Knowing the sequence of nucleotides in the genome of a species will give us information about its genes and their products. This information can be significantly useful in understanding the functioning of the organism. Therefore a heavy emphasis has recently been placed in discovering genomes of species which are important to humans in one way or the other. The most important of them all would off course be us.
Sequencing a genome, i.e., determining the sequence of nucleotides of the genes it comprises of, is not an easy task by any standards because of the enormity of the data volume. For example, the human genome is comprised of over 3 billon letters. Just to give you an idea of the enormity of the information, imagine putting each and every nucleotide of the human genome in a single cell from end to end. It will be about 3 m long. Now multiply it with an average number of cells in a human body, i.e. 1013. Guess how long would that be? The total length will be equal to a return journey between earth and the sun. Staggering! It is a feat of modern Molecular Biology to have completely sequenced the whole human genome. This has been made possible due to recent advances in the automation of sequencing technology. Concurrently, genome projects of several other animal, plant and bacterial species have also been completed while many others are currently underway. The relevant information about the human and other genome projects can be found at the following websites:
http://www.hugo-international.org/
http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml
Definition of Bioinformatics
Modern developments in the laboratory techniques have resulted in high throughput automated methodologies in the areas of DNA sequencing and gene expression studies consequently resulting in to an exponentially ever increasing data. To be of some use the enormous volume of these data has to be sorted and stored in a readily retrievable form as well as be analyzed for eventual interpretation. This challenge cannot be met without using high speed and efficient computer hardware and advance algorithms and software.
In the physical sciences area, Computer Science has excelled more than many other disciplines. Using computers we can deal with the ever increasing volume and kinds of data being accumulated at a speed which was not possible in the past. Computer hardware and software are not only appropriate but have now become essential tools to deal with the kind of data being produced in Molecular Biology. Sequences of these molecules (DNA & proteins) being discovered on an unprecedented speed and scale are stored in large data bases. New software has also been developed to sort, store and analyze these data. Many of these data bases as well as the relevant software are generally available, free of cost, online through the Internet to the public. Computers are currently being used to deal with all sorts of biological data, whether it originates from a population genetics study or comprises of DNA sequences. So would that all be considered under the realm of Bioinformatics? Probably not! In fact, using computer technology to deal with general biological data would be considered under a wider term of computational Biology. While, Bioinformatics is the use of computers to deal particularly with Molecular Biological data, comprising of DNA/RNA and protein sequences as well as gene expression. Briefly, the use of computers in studying genes, their sequences and expression is called Bioinformatics and as such falls under the wider umbrella of computational biology.
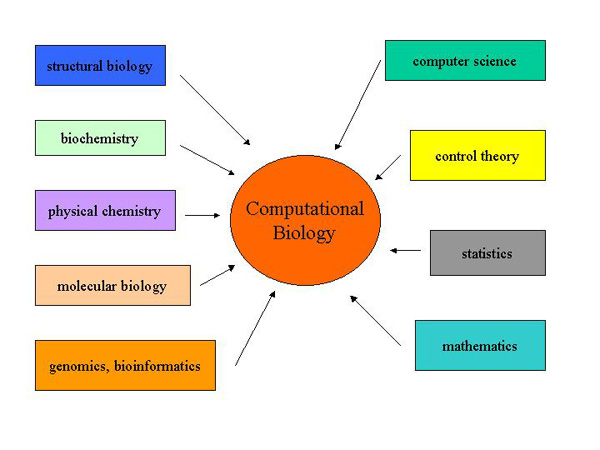
Two Myths about Bioinformatics
Unrealistic expectations have always been attached to new technologies emerging in the past. Not long ago, when genetic engineering was new, people thought that the solution to all biological problems was almost at hand, however, the limitations of the technology, were realized only later despite its enormous theoretical potentials. The other myth attached to most new technologies is that they are too difficult to learn and therefore not every body’s ball game. Both these myths are somehow attached to Bioinformatics and both of them are wrong.
Bioinformatics as a New Utility
Bioinformatics is not a replacement for any existing branch of science. In fact it is just another utility to facilitate scientific research in the areas of life sciences. The biological research carried out in a conventional laboratory is termed as study in vitro literally meaning in glass. Relating to the silicon chips used in computer processors, Bioinformatics has also been termed as study in silico as opposed to study in vitro. Several experiments can now be done in silico using bioinformatics either verifying or altogether replacing the time consuming and expensive in vitro experiments. That is not all. Bioinformatics has also introduced several novel approaches in biological research. Biological sequence similarity searching and sequence based Phylogenetic analysis to study evolutionary relationships among the species are two of the popular examples.
Bioinformatics is Not Rocket Science!
The term “Rocket Science” is generally used for science and technology perceived incomprehensible. Bioinformatics, for sure, is not rocket science. In fact even rocket science itself is not “Rocket Science”. You can comprehend and make use of it if you are determined to do so. However, there is a prevalent hesitation among biologists for using Bioinformatics in their studies. This hesitation is based upon a common misconception that you have to be Computer Scientist to be able to use Bioinformatics. You don’t! A reasonable level of computer literacy is sufficient to be able to make use of Bioinformatics. Most of the biologists have this level of literacy. However, if you want to dwell deeper in to Bioinformatics and want to become a hard core Bioinformatician than you need to have a deeper understanding of programming or system administration skills. For the rest of the biologists, we are already Bioinformatics ready.
Hot Topics in Bioinformatics
Searching for Biological Sequence Similarity
A similarity in structures indicates a similarity in functions. This principle has widely been used through out the evolutionary history of mankind. That is the bases and reason of classification of knowledge of structures and functions of things around us. The same is true for biological molecules like DNA and proteins. If the sequence of a novel protein, for example, is the same as the one already known, so would be its function. Therefore when a new sequence is discovered, it is searched first for similarity in the huge databases of already known sequences. Similarity searching of biological sequences is therefore the most widely used technique in Bioinformatics and the most popular program used for this purpose is the Basic Local Alignment Search Tool or BLAST for short. The following website can provide excellent tutorials and the actual utility itself which is freely available to all users: http://www.ncbi.nlm.nih.gov/blast/
Comparative Genomics
Genomes of several species have already been sequenced and many more projects are currently underway. Information obtained from these projects will provide the basis for a new approach in Biology called comparative genomics similar to the one applied by Charles Darwin. The only difference is that comparative genomics is based on sequences rather than morphology (structure of body parts). Comparing genomes of related species would provide tremendous amount of knowledge of the way genomes are organized and how major evolutionary changes do take place. More information on comparative genomics can be found at the following website: http://www.genome.gov/11509542
Single Nucleotide Polymorphisms (SNPs)
The sequences of DNA and therefore proteins are not different only between species. Individuals of the same species also have small differences among themselves, in fine details. That is the bases for every individual to be different from any other individual. The scientists have noticed that same gene in individuals within a species could exist in several different forms of sequences termed polymorphism. Sometimes the differences could be only at a single nucleotide level, termed Single Nucleotide Polymorphism or SNP (pronounced as snip) for short. Genomes are generally full of SNPs. Most of the SNPs have no direct impact but many others can explain different biological phenomenon.
Let us take an example of a clinical trial of a drug which proves very successful in its action by curing 80% of the patients it is tried on. But what happened with the rest of the 20% who did not respond to such a successful drug? If we investigate deeper we might find that some of the patients not responding to the drug have SNPs in the genes specific for responding to that particular drug. Therefore, by measuring sets of SNPs in multitude of individuals and correlating those to the incidence of disease and variation in drug response one can eventually help in designing novel drugs as well as modifying the existing ones. Moreover, the overall presence of SNPs in the genome also makes them useful markers for other studies. Large scale studies to collect SNP data in human population is a hot topic in today’s Bioinformatics. More information on SNPs can be found at the following website: http://www.ornl.gov/sci/techresources/Human_Genome/faq/snps.shtml
DNA Microarrays
Genes are expressed as proteins and that is how they carry out and sustain life processes in a cell and eventually the organism. Studying gene expression has traditionally been a time consuming exercise because of slow conventional methods and a large number of genes required to be studied before making a generalization. New advances in this field have made such studies exponentially faster. Now, studying expression of hundreds or even thousands of genes simultaneously has become possible with a new technology called DNA Microarrays. In this technique, thousands of genes can be spotted on a small silicon chip and the experimental results are read using sophisticated laser technology and analyzed using advanced software.
Such studies help us identify the genes which are expressed and those which are turned off in certain instances; for example, a normal breast tissue versus a breast tumor. In this example, the proteins expressed exclusively in the breast tumor are potential targets for cancer treatments, vaccines, and other such therapeutics. More information on DNA Microarrays can be found at the following website: http://www.ncbi.nlm.nih.gov/About/primer/microarrays.html
Structural Biology
Structures and functions are strongly correlated in Biology. If we can predict the structure of a protein molecule, we can also predict its function which is a significantly important piece of information in biological research. Studying and predicting 3-D structures of biomolecules like proteins is an integral part of Bioinformatics and is traditionally done by using X-ray Crystallography and Nuclear Magnetic Resonance (NMR). There are great logistic difficulties in this field and therefore very limited information has been made available so far using these traditional methods. Predicting the structures using Bioinformatics techniques could be a more plausible alternative for this purpose. By predicting the structures correctly, we should be able to study the dynamics of its molecular motion, and the specific interactions with drugs and other proteins.
This field can also be very useful in discovering and designing new drugs. An enormous portion of the funds allocated for research and development by multinational pharmaceutical giants is being diverted for discovering more efficient methods in this field. More information on 3-D protein structures can be found at the following website: http://www.pdb.org/pdbstatic/tutorials/tutorial.html
Skills Required For Becoming a Bioinformatician
Bioinformatics is an interaction of more than one disciplines of science and therefore it is necessary to have exposure to them all to be successful in it. Since Bioinformatics deals with Molecular Biological information, it is important to know Molecular Biology first. Since this information is stored away in huge data bases so an understanding of statistics would be helpful, too. Last but not least we need to design and use software using high-powered machines. Therefore an understanding of mathematics and Computer Science on the whole is also important. To put it briefly we need to be well grounded in one of the following subjects and have a working knowledge of the other three:
Career Paths, Users vs. Developers
The Bioinformatic tools are developed by professionals who are expert in Computer Sciences having some understanding of life sciences and mainly used by Molecular Biologists. Therefore there are two alternative career paths in this field. If you are a Molecular Biologist, you can become a user by learning the use of various Bioinformatics resources. If, however, you find Bioinformatics interesting enough, you can also develop your skills in programming and become a developer in the field. For Computer Scientists becoming a developer could be a more appropriate choice after learning the basics of the relevant subjects like Molecular Biology. They can make a career out of Bioinformatics either as a system administrator or a program developer. As a system administrator you setup and manage Bioinformatic Servers providing various services used in Bioinformatics, like large sequence data bases and suits of software for analysis of such data. As a developer, you design new data bases and write novel software using various programming languages.
Learning Bioinformatics through Self Education
Because this is a new field, it is hard to find people who are formally qualified in this area. Most of the Bioinformaticians today are self-educated professionals. They are either the Computer Scientist who have developed an understanding of Molecular Biology or the Molecular Biologists who have learnt Computer Science. Either way these professionals are currently in high demand. The introduction of Bioinformatics in to Universities can be related to the introduction of computers which happened in not too distant past. Most of us have seen it happening in our life time. At that time there were not many formally qualified Computer Scientists. In fact not many Universities were even offering any formal qualification in this field until then. Most people, therefore, learnt computers through self education and later on excelled in the field according to their particular needs and interests. A similar pattern is now being observed in the field of Bioinformatics. There are not many formally qualified personnel in this field right now; therefore, self education is the most suitable way to learn this new field. The self-learners of Bioinformatics are in much better situation than those for Computer Sciences. A huge learning resource is available through the Internet which was not available at the time the computers were introduced due to obvious reasons. There are many websites which are currently providing excellent learning resources in this area. One such site is: www.ebi.ac.uk/2can/home.html
Moreover, there are many good books in the print form as well as E-books to help the self learners. Attending workshops on Bioinformatics offered sporadically in Pakistan can also be a quick and effective way of learning this subject through hands on practice sessions offered in these workshops. One such workshop was organized by the authors under the umbrella of COMSTECH recently.
Learning Bioinformatics through Formal Education
Bioinformatics can be a rewarding career for students continuing for their undergraduate studies. Besides COMSATS University (CU), two other Universities, i.e.; the Mohammad Ali Jinah University and the International Islamic University, are currently offering undergraduate degree programs in Bioinformatics in Islamabad. At a graduate level, the field of Bioinformatics is open both for Biologists and Computer Scientists alike. Postgraduate studies in the field could be an option for both of them, although, a postgraduate degree is not necessary to get you started in the new field. Rather, a certificate or a diploma should be sufficient for this purpose.
Currently no University in Pakistan that the authors know of is offering any postgraduate studies in Bioinformatics and therefore the interested graduates have to look elsewhere. Besides traveling to overseas they can also do postgraduate certificate or diploma courses through correspondence as offered by a few Universities internationally. Such courses are logistically and financially more feasible and usually equally recognized. Various international Universities offer specific streams of studies both for Biology and computer graduates. Searching on the Internet may lead you to a few places of your interest.
The Scope of Bioinformatics
Bioinformatics is a newly emerging field worldwide and more so in Pakistan. The exponentially growing huge volumes of sequence and expression data need a large number of skilled personnel in the field. Many multinational pharmaceutical companies are investing billions of dollars in research and development. A significant portion of this money is diverted to design and discover new drugs with the help of Bioinformatics tools. Unfortunately, no such initiative is currently underway in Pakistan. Like many other technologies, Bioinformatics is yet far from becoming an industry in Pakistan. However, it does not necessarily mean that there is no scope for Bioinformatics professional in Pakistan. There is a growing scope for Bioinformatics trained personnel in Pakistan in the public sector. The Higher Education Commission of Pakistan has declared Bioinformatics a mandatory part of curriculum for all degree programs of Biology. That initiative alone will enhance significantly the demand of fresh graduates in Pakistan. Initiatives are also underway to setup centers of excellence in Bioinformatics where new professionals will be trained and Bioinformatics servers will be established. That would be another possible market for the local graduates. With these initiatives and many more to come, Pakistan will provide a sizeable market for Bioinformatics graduates in the fields of research and teaching within the country for some time to come.
The CU Promoting Bioinformatics in Pakistan
The CU is at the forefront of promoting Bioinformatics in Pakistan. This is the first institute in the public sector to introduce a formal degree in Bioinformatics. The authors have also organized a couple of major international workshops in the area. The recent one was organized at the platform of COMSTECH during August & September where participants and the instructors came from twelve different countries. The CU is now in the process of organizing a series of bi-monthly workshops for the researchers and students from all parts of Pakistan.
The CU has also recently been elected Pakistan National Node of EMBnet (www.embnet.org) which is a premier international organization in the area of Bioinformatics. This alliance has provided CU specifically and Pakistan in general, access to the multitude of international experts in the area. Raheel is the national node manager and Shahid is staff member. The objective of the node is to promote education and training as well as providing local facilities in the field of Bioinformatics within the country. The first facility of this kind has already been set up in the form of a Bioinformatics server providing a suit of software to be used for Bioinformatics analyses. We are currently in the process of upgrading this server with a relatively more powerful machine enabling a larger number of users to login simultaneously. The present server can be accessed at the following web address:
http://mail.comsats.edu.pk/mail/bioinformatics.htm
[Shahid is a HEC foreign faculty in the department of Biosciences at the COMSATS University, Islamabad (Email: snchohan@hotmail.com)
Raheel is Professor and Chairman at the department of Biosciences at the COMSATS University, Islamabad (Email: raheelqamar@hotmail.com)
http://www.hec.gov.pk/htmls/news_letter/issues/2006/November_06.pdf